Welcome to the
Abu-Remaileh Lab at Stanford University
We study the molecular and cellular bases of metabolic adaptation in health and disease
OUR RESEARCH
We are interested in identifying novel pathways that enable cellular and organismal adaptation to metabolic stress and changes in environmental conditions. We also study how these pathways go awry in human diseases such as cancer, neurodegeneration and metabolic syndrome, in order to engineer new therapeutic modalities.
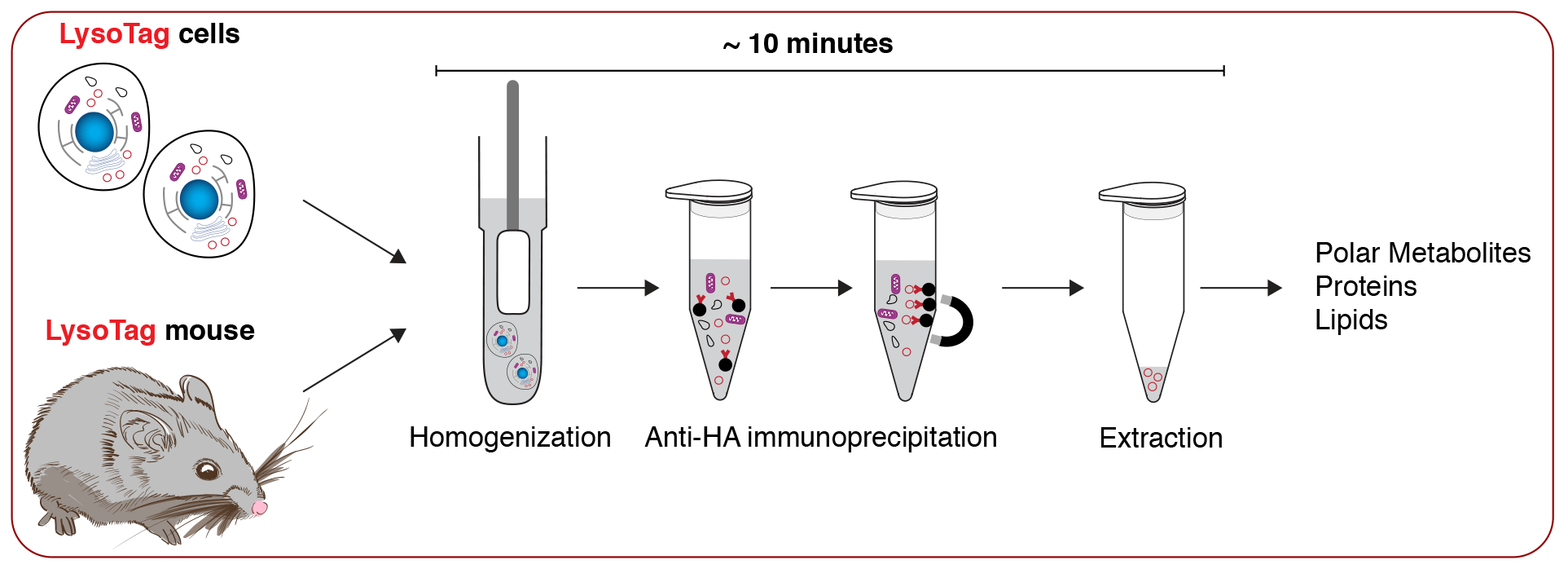
To address these questions, our lab uses a multidisciplinary approach to study the biochemical functions of the lysosome in vitro and in vivo. Lysosomes are membrane-bound compartments that degrade macromolecules and clear damaged organelles to enable cellular adaptation to various metabolic states. Lysosomal function is critical for organismal homeostasis—mutations in genes encoding lysosomal proteins cause severe human disorders known as lysosomal storage diseases, and lysosome dysfunction is implicated in age-associated diseases including cancer, neurodegeneration and metabolic syndrome.

By developing novel tools and harnessing the power of metabolomics, proteomics and functional genomics, our lab will define 1) how the lysosome communicates with other cellular compartments to fulfill the metabolic demands of the cell under various metabolic states, 2) and how its dysfunction leads to rare and common human diseases. Using insights from our research, we will engineer novel therapies to modulate the pathways that govern human disease.
PEOPLE

Monther Abu-Remaileh, Ph.D.
ChEM-H Institute Scholar
Assistant Professor of Chemical Engineering
Assistant Professor of Genetics
Email: monther (at) stanford.edu

Aleksandra Levina, Ph.D.
Postdoctoral fellow
Email: alevina (at) stanford.edu

Wentao Dong, Ph.D.
Postdoctoral fellow
Email: wdong5 (at) stanford.edu

Ali Ghoochani, Ph.D.
Postdoctoral fellow
Email: aligh (at) stanford.edu

Cindy Lin
Grad Student/Genetics
Email: clin5 (at) stanford.edu


Austin Murchison
Grad Student/Biology
Email: austinmu (at) stanford.edu


Uche Medoh
Grad Student/Biochemistry
Email: uchemed (at) stanford.edu

Julia Heiby, Ph.D.
Joint postdoctoral fellow with Dr. Alessandro Ori/FLI
Email: Julia.Heiby (at) leibniz-fli.de

Jian Xiong
Postdoctoral fellow
Email: xiongj (at) stanford.edu
Samantha Scharenberg
Grad Student/MD-PhD/Biophysics
Email: sscharen (at) stanford.edu


George Walters-Marrah
Grad Student/Biophysics
Email: gwmarrah (at) stanford.edu
Kwamina Nyame
Grad Student/Biochemistry
Email: kwaminanyame (at) stanford.edu
Andy Hims
research assistant
Email: ahims (at) stanford.edu



Hisham Alsohybe
Grad Student/Biophysics
Email: hishama (at) stanford.edu
Vincent Hernandez III
research assistant
Email: vhernan3 (at) stanford.edu
Nickolas Manfred
Grad Student/Neuroscience
Email: nickmanfred (at) stanford.edu
SELECTED PUBLICATIONS
1. Medoh UN, Hims A, Chen JY, Ghoochani A, Nyame K, Dong W, Abu-Remaileh M. The Batten disease gene product CLN5 is the lysosomal bis(monoacylglycero)phosphate synthase. Science. 2023 Sep 15;381(6663):1182-1189. doi: 10.1126/science.adg9288.
2. Fasimoye R, Dong W, Nirujogi RS, Rawat ES, Iguchi M, Nyame K, Phung TK, Bagnoli E, Prescott AR, Alessi DR#, Abu-Remaileh M#. Golgi-IP, a tool for multimodal analysis of Golgi molecular content. PNAS. 2023 May 16;120(20):e2219953120. doi: 10.1073/pnas.2219953120.
3. Scharenberg SG, Dong W, Ghoochani A, Nyame K, Levin-Konigsberg R, Krishnan AR, Rawat ES, Spees K, Bassik MC, Abu-Remaileh M. An SPNS1-dependent lysosomal lipid transport pathway that enables cell survival under choline limitation. Science Advances. 2023 Apr 21;9(16):eadf8966. doi: 10.1126/sciadv.adf8966.
4. Laqtom NN, Dong W, Medoh UN, Cangelosi AL, Dharamdasani V, Chan SH, Kunchok T, Lewis CA, Heinze I, Tang R, Grimm C, Dang Do AN, Porter FD, Ori A, Sabatini DM, Abu-Remaileh M. CLN3 is required for the clearance of glycerophosphodiesters from lysosomes. Nature. 2022 Sep;609(7929):1005-1011. doi: 10.1038/s41586-022-05221-y.
5. Armenta DA, Laqtom NN, Alchemy G, Dong W, Morrow D, Poltorack CD, Nathanson DA, Abu-Remalieh M#, Dixon SJ#. Ferroptosis inhibition by lysosome-dependent catabolism of extracellular protein. Cell Chem Biol. 2022 Nov 17;29(11):1588-1600.e7. doi: 10.1016/j.chembiol.2022.10.006. (#co-corrsponding author)
6. Crossley MP, Song C, Bocek MJ, Choi JH, Kousorous J, Sathirachinda A, Lin C, Brickner JR, Bai G, Lans H, Vermeulen W, Abu-Remaileh M, Cimprich KA. R-loop-derived cytoplasmic RNA-DNA hybrids activate an immune response. Nature. 2023 Jan;613(7942):187-194. doi: 10.1038/s41586-022-05545-9.
7. Pedram K, Laqtom NN, Shon DJ, Di Spiezio A, Riley NM, Saftig P, Abu-Remaileh M, Bertozzi CR. Lysosomal cathepsin D mediates endogenous mucin glycodomain catabolism in mammals. PNAS. 2022 Sep 27;119(39):e2117105119. doi: 10.1073/pnas.2117105119.
8. Dong W, Rawat ES, Stephanopoulos G#, Abu-Remaileh M#. Isotope tracing in health and disease. Curr Opin Biotechnol. 2022 Aug;76:102739. doi: 10.1016/j.copbio.2022.102739. Epub 2022 Jun 20. Review.
9. Medoh UN, Chen JY, Abu-Remaileh M. Lessons from metabolic perturbations in lysosomal storage disorders for neurodegeneration. Curr Opin Sys Bio. 2022 March; https://doi.org/10.1016/j.coisb.2021.100408.
For full list visti Google Scholar.

JOIN US!
Graduate Students:
Current Stanford graduate students from all programs (PhD, MD, PhD/MD) who are interested in joining our lab are welcome to contact Monther directly. Please send a current CV in pdf format.
Postdocs and Clinical Fellows:
Motivated postdocs and clinical fellows with expertise in biochemistry, molecular biology, metabolism, mouse physiology or neurobiology are encouraged to apply. Please email a cover letter, your CV, and contact information for three references to Monther.
Undergradaute Students:
Students who are interested in conducting biomedical research and being part of our multidisciplinary team are welcome to email Monther for opportunities.
Master Students:
Current masters students who are enrolled in national or international programs and hoping to join our lab as visiting students (6-12 month duration) to complete their thesis are welcome. Research experience in biology, biochemistry or chemistry is an advantage. Please email a cover letter, your CV, and contacts of two references to Monther.
CONTACT US!
Address:
ChEM-H Building, Room: N227
290 Jane Stanford Way
Stanford, CA 94305

Our Lab is located at the new building that hosts both ChEM-H and Wu Tsai Neuroscience Research Center.